Estimating effect of multiple treatments#
[1]:
from dowhy import CausalModel
import dowhy.datasets
import warnings
warnings.filterwarnings('ignore')
[2]:
data = dowhy.datasets.linear_dataset(10, num_common_causes=4, num_samples=10000,
num_instruments=0, num_effect_modifiers=2,
num_treatments=2,
treatment_is_binary=False,
num_discrete_common_causes=2,
num_discrete_effect_modifiers=0,
one_hot_encode=False)
df=data['df']
df.head()
[2]:
| X0 | X1 | W0 | W1 | W2 | W3 | v0 | v1 | y | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.782955 | -0.117877 | 0.556503 | 0.576597 | 0 | 2 | 10.075403 | 5.888747 | 2.584256 |
| 1 | 0.533871 | 0.225779 | 1.091080 | 0.106325 | 2 | 2 | 10.368336 | 10.836949 | 496.136374 |
| 2 | 0.719629 | 0.823192 | -0.619869 | 0.614200 | 1 | 1 | 4.088105 | 7.815383 | 300.441897 |
| 3 | -0.596388 | -0.462312 | 0.304888 | 0.076668 | 3 | 2 | 7.784805 | 14.458986 | -184.907269 |
| 4 | 1.470431 | 1.013644 | 0.009036 | -2.402029 | 2 | 3 | 1.394636 | 1.845187 | 55.330271 |
[3]:
model = CausalModel(data=data["df"],
treatment=data["treatment_name"], outcome=data["outcome_name"],
graph=data["gml_graph"])
[4]:
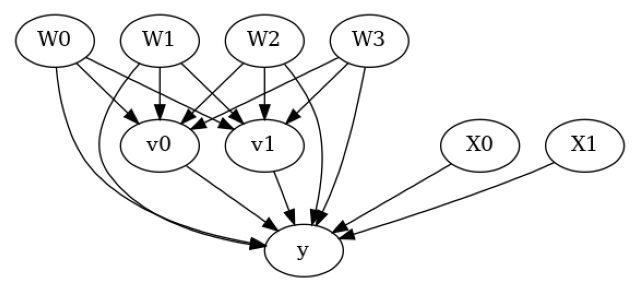
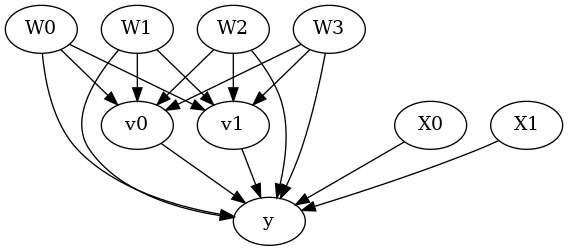
model.view_model()
from IPython.display import Image, display
display(Image(filename="causal_model.png"))
[5]:
identified_estimand= model.identify_effect(proceed_when_unidentifiable=True)
print(identified_estimand)
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
─────────(E[y|W2,W0,W1,W3])
d[v₀ v₁]
Estimand assumption 1, Unconfoundedness: If U→{v0,v1} and U→y then P(y|v0,v1,W2,W0,W1,W3,U) = P(y|v0,v1,W2,W0,W1,W3)
### Estimand : 2
Estimand name: iv
No such variable(s) found!
### Estimand : 3
Estimand name: frontdoor
No such variable(s) found!
Linear model#
Let us first see an example for a linear model. The control_value and treatment_value can be provided as a tuple/list when the treatment is multi-dimensional.
The interpretation is change in y when v0 and v1 are changed from (0,0) to (1,1).
[6]:
linear_estimate = model.estimate_effect(identified_estimand,
method_name="backdoor.linear_regression",
control_value=(0,0),
treatment_value=(1,1),
method_params={'need_conditional_estimates': False})
print(linear_estimate)
*** Causal Estimate ***
## Identified estimand
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
─────────(E[y|W2,W0,W1,W3])
d[v₀ v₁]
Estimand assumption 1, Unconfoundedness: If U→{v0,v1} and U→y then P(y|v0,v1,W2,W0,W1,W3,U) = P(y|v0,v1,W2,W0,W1,W3)
## Realized estimand
b: y~v0+v1+W2+W0+W1+W3+v0*X0+v0*X1+v1*X0+v1*X1
Target units: ate
## Estimate
Mean value: 26.349570350104635
You can estimate conditional effects, based on effect modifiers.
[7]:
linear_estimate = model.estimate_effect(identified_estimand,
method_name="backdoor.linear_regression",
control_value=(0,0),
treatment_value=(1,1))
print(linear_estimate)
*** Causal Estimate ***
## Identified estimand
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
─────────(E[y|W2,W0,W1,W3])
d[v₀ v₁]
Estimand assumption 1, Unconfoundedness: If U→{v0,v1} and U→y then P(y|v0,v1,W2,W0,W1,W3,U) = P(y|v0,v1,W2,W0,W1,W3)
## Realized estimand
b: y~v0+v1+W2+W0+W1+W3+v0*X0+v0*X1+v1*X0+v1*X1
Target units:
## Estimate
Mean value: 26.349570350104635
### Conditional Estimates
__categorical__X0 __categorical__X1
(-4.113, -0.663] (-3.479, -0.833] -97.708112
(-0.833, -0.255] -52.276502
(-0.255, 0.251] -22.606347
(0.251, 0.847] 7.788871
(0.847, 4.047] 54.294575
(-0.663, -0.0794] (-3.479, -0.833] -65.348499
(-0.833, -0.255] -21.277903
(-0.255, 0.251] 7.985988
(0.251, 0.847] 35.896265
(0.847, 4.047] 83.248394
(-0.0794, 0.414] (-3.479, -0.833] -49.649198
(-0.833, -0.255] -2.170874
(-0.255, 0.251] 26.187706
(0.251, 0.847] 55.797786
(0.847, 4.047] 100.403169
(0.414, 1.005] (-3.479, -0.833] -30.638770
(-0.833, -0.255] 15.790545
(-0.255, 0.251] 43.731940
(0.251, 0.847] 73.447815
(0.847, 4.047] 120.114812
(1.005, 4.298] (-3.479, -0.833] -1.009736
(-0.833, -0.255] 46.841476
(-0.255, 0.251] 75.184308
(0.251, 0.847] 103.445271
(0.847, 4.047] 151.322717
dtype: float64
More methods#
You can also use methods from EconML or CausalML libraries that support multiple treatments. You can look at examples from the conditional effect notebook: https://py-why.github.io/dowhy/example_notebooks/dowhy-conditional-treatment-effects.html
Propensity-based methods do not support multiple treatments currently.