Demo for the DoWhy causal API#
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
from dowhy.graph import build_graph_from_str
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
nx_graph = build_graph_from_str(data["dot_graph"])
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | 0.319949 | True | 5.882584 |
| 1 | 0.440958 | True | 4.622104 |
| 2 | -0.880456 | False | 0.734731 |
| 3 | 1.352040 | False | 2.243402 |
| 4 | -0.119246 | True | 5.176788 |
| ... | ... | ... | ... |
| 995 | 0.223470 | False | 1.116045 |
| 996 | 0.450676 | True | 6.721430 |
| 997 | 0.814874 | True | 4.027162 |
| 998 | 1.359336 | True | 7.286949 |
| 999 | -0.515406 | False | 0.779886 |
1000 rows × 3 columns
[3]:
# data['df'] is just a regular pandas.DataFrame
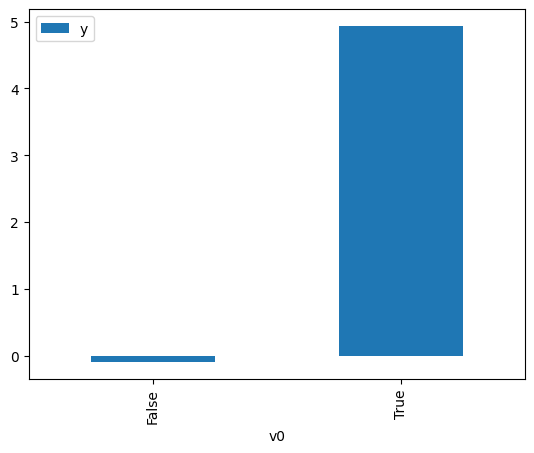
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<Axes: xlabel='v0'>
[4]:
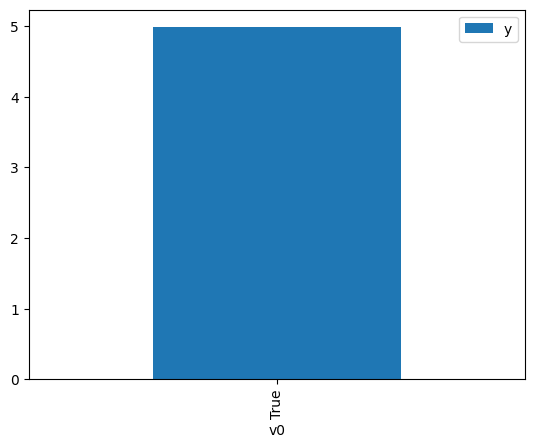
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause]
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<Axes: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 0.259164 | False | -1.025702 | 0.499463 | 2.002150 |
| 1 | 0.330791 | False | 1.201951 | 0.492570 | 2.030169 |
| 2 | 0.289888 | False | 0.844696 | 0.496506 | 2.014074 |
| 3 | 1.543579 | False | 0.812667 | 0.378333 | 2.643175 |
| 4 | 0.037070 | False | 0.141225 | 0.520827 | 1.920024 |
| ... | ... | ... | ... | ... | ... |
| 995 | -0.448824 | False | -0.357686 | 0.567196 | 1.763060 |
| 996 | 0.934334 | False | 2.766019 | 0.434853 | 2.299629 |
| 997 | -0.348975 | False | 0.473715 | 0.557736 | 1.792964 |
| 998 | 0.821882 | False | 0.798649 | 0.445521 | 2.244566 |
| 999 | 0.287035 | False | 0.391905 | 0.496781 | 2.012961 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 3.516035 | True | 9.864709 | 0.778337 | 1.284791 |
| 1 | -0.783727 | True | 3.441380 | 0.401468 | 2.490858 |
| 2 | -0.559160 | True | 1.803474 | 0.422408 | 2.367380 |
| 3 | -0.010100 | True | 4.910736 | 0.474643 | 2.106847 |
| 4 | 1.094514 | True | 4.930522 | 0.580237 | 1.723434 |
| ... | ... | ... | ... | ... | ... |
| 995 | 1.607544 | True | 5.726897 | 0.627441 | 1.593774 |
| 996 | 0.237660 | True | 5.249902 | 0.498467 | 2.006150 |
| 997 | -0.226316 | True | 4.621072 | 0.453942 | 2.202925 |
| 998 | -1.045048 | True | 4.631207 | 0.377551 | 2.648650 |
| 999 | -0.789050 | True | 5.500858 | 0.400976 | 2.493916 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression#
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 4.80317963672906$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.136134908742183$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.947 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.947 |
| Method: | Least Squares | F-statistic: | 8904. |
| Date: | Thu, 12 Feb 2026 | Prob (F-statistic): | 0.00 |
| Time: | 23:22:09 | Log-Likelihood: | -1459.8 |
| No. Observations: | 1000 | AIC: | 2924. |
| Df Residuals: | 998 | BIC: | 2933. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 1.0730 | 0.031 | 34.403 | 0.000 | 1.012 | 1.134 |
| x2 | 4.9654 | 0.052 | 96.222 | 0.000 | 4.864 | 5.067 |
| Omnibus: | 0.793 | Durbin-Watson: | 2.074 |
|---|---|---|---|
| Prob(Omnibus): | 0.673 | Jarque-Bera (JB): | 0.684 |
| Skew: | -0.054 | Prob(JB): | 0.711 |
| Kurtosis: | 3.068 | Cond. No. | 2.11 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.