Demo for the DoWhy causal API#
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
from dowhy.graph import build_graph_from_str
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
nx_graph = build_graph_from_str(data["dot_graph"])
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | 0.266331 | False | 0.186198 |
| 1 | -0.577283 | False | -0.263693 |
| 2 | -0.759739 | True | 3.956742 |
| 3 | -0.158249 | False | 0.146362 |
| 4 | 0.129720 | False | -0.781201 |
| ... | ... | ... | ... |
| 995 | 0.129913 | False | -1.043541 |
| 996 | -1.596959 | False | -2.221180 |
| 997 | -0.573585 | False | -0.898515 |
| 998 | -1.221957 | False | -2.245137 |
| 999 | 0.132693 | False | 0.425453 |
1000 rows × 3 columns
[3]:
# data['df'] is just a regular pandas.DataFrame
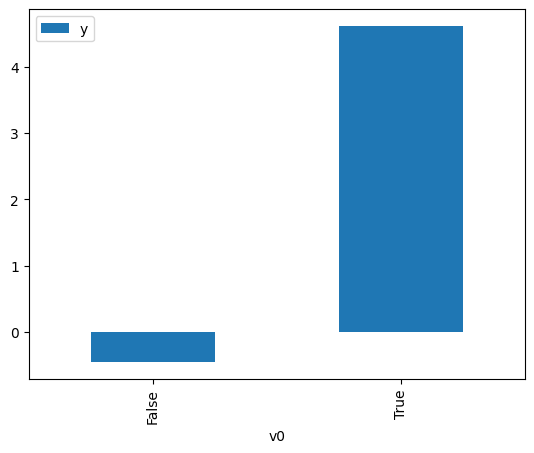
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<Axes: xlabel='v0'>
[4]:
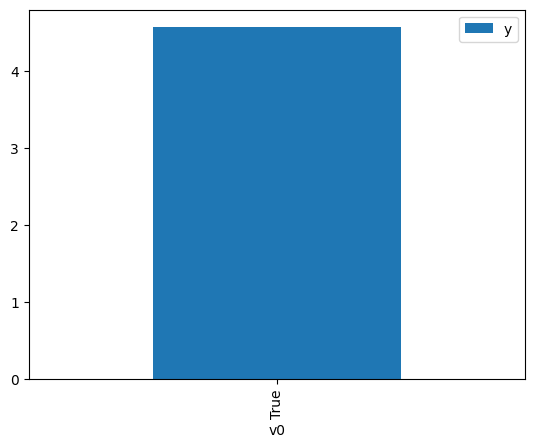
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause]
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<Axes: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -1.461199 | False | -0.974013 | 0.859671 | 1.163236 |
| 1 | -1.183711 | False | -0.711176 | 0.812896 | 1.230169 |
| 2 | -2.022528 | False | -3.317512 | 0.924678 | 1.081458 |
| 3 | 1.225780 | False | 2.189129 | 0.180229 | 5.548483 |
| 4 | -0.393947 | False | -3.161741 | 0.620331 | 1.612041 |
| ... | ... | ... | ... | ... | ... |
| 995 | -0.707376 | False | 0.270499 | 0.706632 | 1.415164 |
| 996 | -0.728482 | False | -0.762697 | 0.712021 | 1.404453 |
| 997 | 0.272532 | False | 1.806332 | 0.417180 | 2.397049 |
| 998 | -1.728074 | False | -0.945565 | 0.895017 | 1.117298 |
| 999 | -0.797739 | False | 0.124563 | 0.729282 | 1.371212 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 0.160562 | True | 3.561945 | 0.548771 | 1.822254 |
| 1 | 0.110464 | True | 4.816448 | 0.533368 | 1.874880 |
| 2 | 0.673347 | True | 6.475563 | 0.696503 | 1.435744 |
| 3 | 0.407568 | True | 6.274436 | 0.622831 | 1.605573 |
| 4 | 0.736094 | True | 4.870072 | 0.712673 | 1.403168 |
| ... | ... | ... | ... | ... | ... |
| 995 | 0.437025 | True | 5.410330 | 0.631361 | 1.583881 |
| 996 | -2.089242 | True | 4.667029 | 0.069766 | 14.333545 |
| 997 | 0.256726 | True | 5.886936 | 0.578054 | 1.729943 |
| 998 | 0.951251 | True | 5.494223 | 0.764018 | 1.308869 |
| 999 | -1.741214 | True | 2.519213 | 0.103464 | 9.665161 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression#
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 5.10681415541703$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.125589635718884$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.928 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.928 |
| Method: | Least Squares | F-statistic: | 6411. |
| Date: | Sat, 08 Nov 2025 | Prob (F-statistic): | 0.00 |
| Time: | 05:28:06 | Log-Likelihood: | -1408.9 |
| No. Observations: | 1000 | AIC: | 2822. |
| Df Residuals: | 998 | BIC: | 2832. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 0.9856 | 0.030 | 32.847 | 0.000 | 0.927 | 1.044 |
| x2 | 5.0376 | 0.049 | 102.816 | 0.000 | 4.941 | 5.134 |
| Omnibus: | 0.277 | Durbin-Watson: | 1.968 |
|---|---|---|---|
| Prob(Omnibus): | 0.871 | Jarque-Bera (JB): | 0.312 |
| Skew: | 0.039 | Prob(JB): | 0.855 |
| Kurtosis: | 2.964 | Cond. No. | 1.67 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.