Note
Click here to download the full example code
Causal discovery with interventional data - Sachs dataset#
We will analyze the Sachs dataset [1] and reproduce analyses
from the Supplemental Figure 8 in [2] demonstrating the
usage of the dodiscover.constraint.PsiFCI algorithm for learning causal graphs
from observational and/or interventional data.
The Sachs dataset is a famous dataset in causal discovery because of its real-life
applicability and access to experimental data that analyzed the causal network of
protein signaling pathways. We will analyze the preprocessed interventional dataset,
which we download using the package pooch.
The preprocessed dataset consists of categorical features, so we will use the
dodiscover.ci.GSquareCITest for testing conditional independence and
invariances of the conditional distributions across experimental conditions.
There are a total of 6 experimental conditions represented by the INT column.
Authors: Adam Li <adam2392@gmail.com>
License: BSD (3-clause)
from pywhy_graphs.viz import draw
from dodiscover.ci import GSquareCITest
from dodiscover import PsiFCI, Context, make_context, InterventionalContextBuilder
import pandas as pd
import bnlearn
import pooch
Pull in the Sachs Dataset#
The Sachs dataset is a famous dataset in causal discovery because of its real-life applicability and access to experimental data that analyzed the causal network of 11 proteins using knockouts and spikings [1]. The pathways for those proteins are already known, so it is an ideal dataset for benchmarking causal discovery algorithms.
We will download a preprocessed version of the dataset from the following url: https://www.bnlearn.com/book-crc/code/sachs.interventional.txt.gz
Ref: https://erdogant.github.io/bnlearn/pages/html/bnlearn.bnlearn.html#bnlearn.bnlearn.import_example # noqa
# use pooch to download robustly from a url
url = "https://www.bnlearn.com/book-crc/code/sachs.interventional.txt.gz"
file_path = pooch.retrieve(
url=url,
known_hash="md5:39ee257f7eeb94cb60e6177cf80c9544",
)
df = pd.read_csv(file_path, delimiter=" ")
# the ground-truth dag is shown here: XXX: comment in when errors are fixed
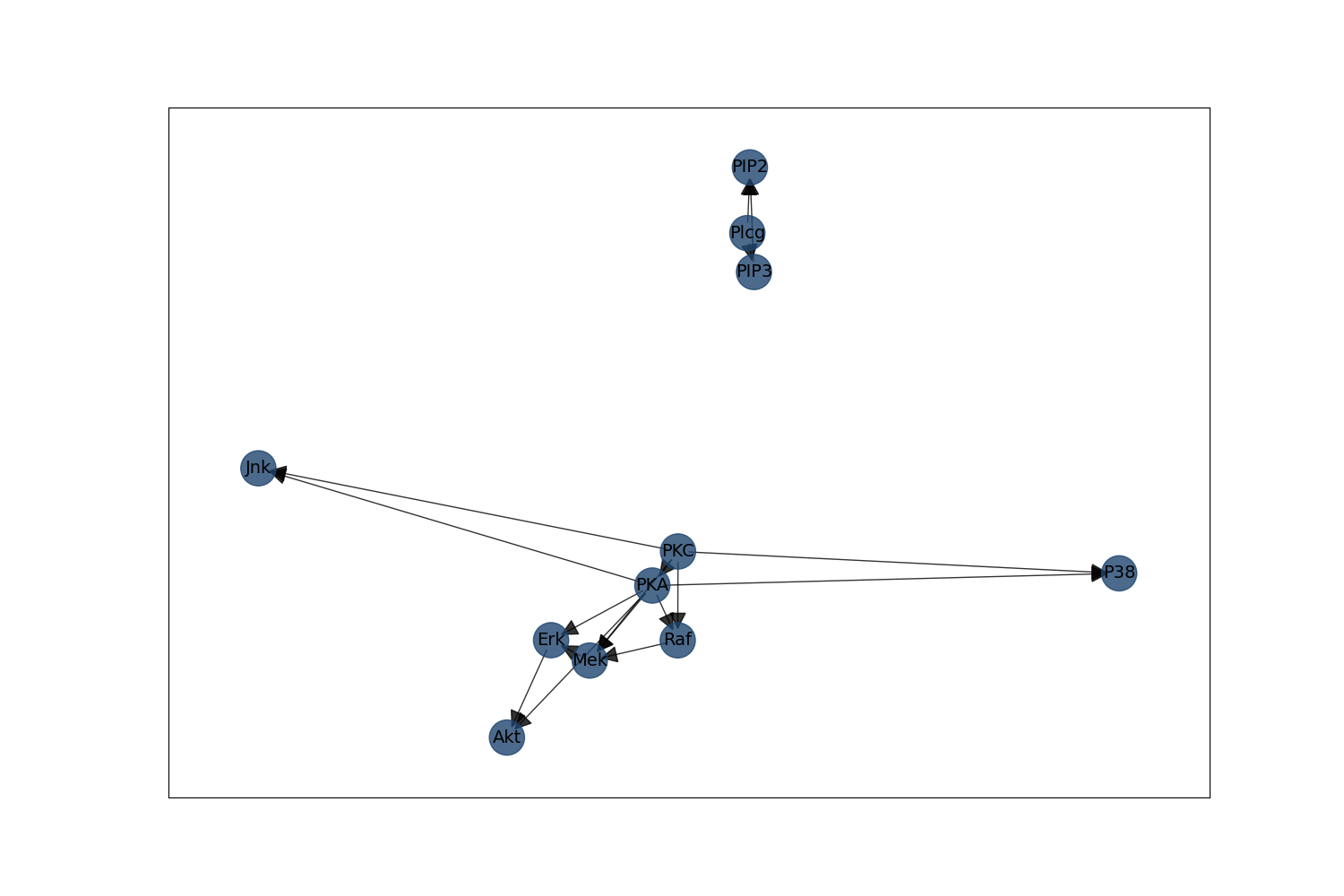
ground_truth_dag = bnlearn.import_DAG("sachs", verbose=False)
fig = bnlearn.plot(ground_truth_dag)
# .. note::
# The Sachs dataset has previously been preprocessed, and the steps are described
# in bnlearn, at the web-page https://www.bnlearn.com/research/sachs05/.
print(df.head())
print(df.shape)
Downloading data from 'https://www.bnlearn.com/book-crc/code/sachs.interventional.txt.gz' to file '/home/circleci/.cache/pooch/08b7ab6b909b20c5ff42bc7d7721556c-sachs.interventional.txt.gz'.
[bnlearn] >Downloading example [sachs] dataset..
[bnlearn] >Set node properties.
[bnlearn] >Set edge properties.
[bnlearn] >Plot based on Bayesian model
Raf ... INT
0 1 ... 8
1 1 ... 8
2 1 ... 8
3 1 ... 8
4 1 ... 8
[5 rows x 12 columns]
(5400, 12)
Preprocess the dataset#
Since the data is one dataframe, we need to process it into a form
that is acceptable by dodiscover’s constraint.PsiFCI algorithm. We
will form a list of separate dataframes.
unique_ints = df["INT"].unique()
# get the list of intervention targets and list of dataframe associated with each intervention
intervention_targets = [df.columns[idx] for idx in unique_ints]
data_cols = [col for col in df.columns if col != "INT"]
data = []
for interv_idx in unique_ints:
_data = df[df["INT"] == interv_idx][data_cols]
data.append(_data)
print(len(data), len(intervention_targets))
6 6
Setup constraint-based learner#
Since we have access to interventional data, the causal discovery algorithm we will use that leverages CI and CD tests to estimate causal constraints is the Psi-FCI algorithm [2].
# Our dataset is comprised of discrete valued data, so we will utilize the
# G^2 (Chi-square) CI test.
ci_estimator = GSquareCITest(data_type="discrete")
# Since our data is entirely discrete, we can also use the G^2 test as our
# CD test.
cd_estimator = GSquareCITest(data_type="discrete")
alpha = 0.05
learner = PsiFCI(
ci_estimator=ci_estimator,
cd_estimator=cd_estimator,
alpha=alpha,
max_combinations=10,
max_cond_set_size=4,
n_jobs=-1,
)
# create context with information about the interventions
ctx_builder = make_context(create_using=InterventionalContextBuilder)
ctx: Context = (
ctx_builder.variables(data=data[0]).num_distributions(6).obs_distribution(False).build()
)
print(ctx.init_graph)
print(ctx.f_nodes)
Graph with 26 nodes and 325 edges
[('F', 0), ('F', 1), ('F', 2), ('F', 3), ('F', 4), ('F', 5), ('F', 6), ('F', 7), ('F', 8), ('F', 9), ('F', 10), ('F', 11), ('F', 12), ('F', 13), ('F', 14)]
Run the learning process#
We have setup our causal context and causal discovery learner, so we will now
run the algorithm using the constraint.PsiFCI.learn_graph() API, which is similar
to scikit-learn’s fit design. All fitted attributes contain an underscore at the end.
learner = learner.learn_graph(data, ctx)
Analyze the results#
Now that we have learned the graph, we will show it here. Note differences and similarities to the ground-truth DAG that is “assumed”. Moreover, note that this reproduces Supplementary Figure 8 in [2].
est_pag = learner.graph_
print(f"There are {len(est_pag.to_undirected().edges)} edges in the resulting PAG")
There are 157 edges in the resulting PAG
Visualize the full graph including the F-node
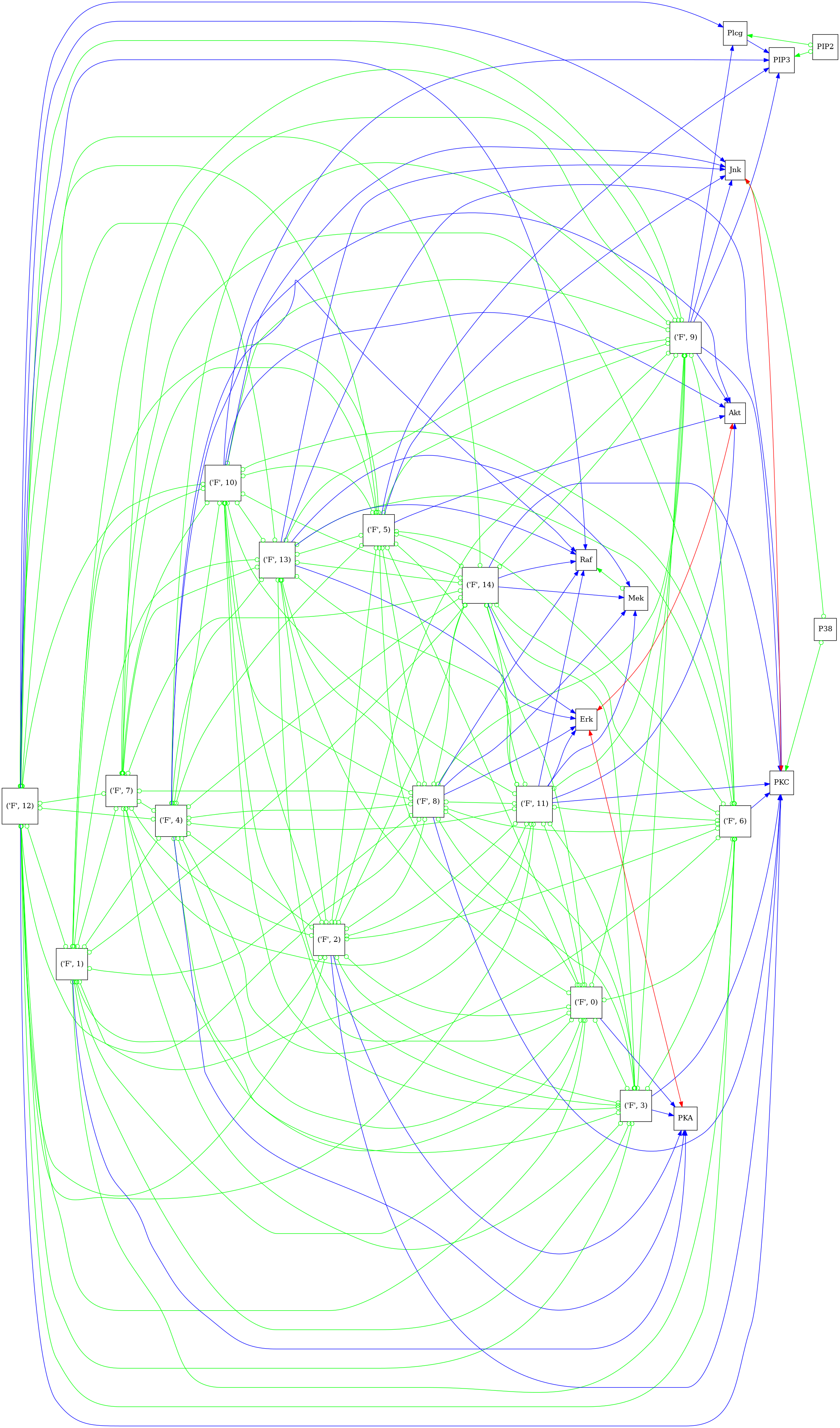
'psi_pag_full.png'
Visualize the graph without the F-nodes
est_pag_no_fnodes = est_pag.subgraph(ctx.get_non_augmented_nodes())
dot_graph = draw(est_pag_no_fnodes, direction="LR")
dot_graph.render(outfile="psi_pag.png", view=True, cleanup=True)
# Interpretation
# --------------
# Looking at the supplemental figure 8b in :footcite:`Jaber2020causal`, we see that the
# learned PAG matches quite well.
# References
# ----------
# .. footbibliography::
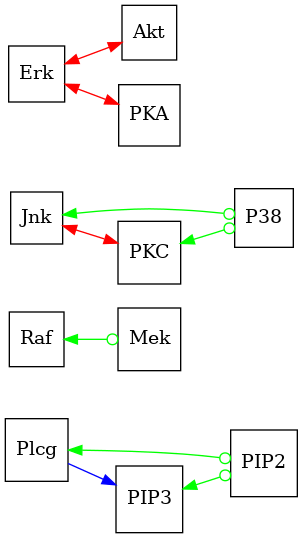
'psi_pag.png'
Total running time of the script: ( 3 minutes 41.070 seconds)