Estimating effect of multiple treatments#
[1]:
from dowhy import CausalModel
import dowhy.datasets
import warnings
warnings.filterwarnings('ignore')
[2]:
data = dowhy.datasets.linear_dataset(10, num_common_causes=4, num_samples=10000,
num_instruments=0, num_effect_modifiers=2,
num_treatments=2,
treatment_is_binary=False,
num_discrete_common_causes=2,
num_discrete_effect_modifiers=0,
one_hot_encode=False)
df=data['df']
df.head()
[2]:
| X0 | X1 | W0 | W1 | W2 | W3 | v0 | v1 | y | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.893067 | 0.103583 | 0.048774 | 0.972218 | 1 | 2 | 13.820447 | 9.255445 | 1131.823214 |
| 1 | 0.513345 | 2.809553 | -0.298139 | 0.897915 | 3 | 2 | 14.397815 | 13.311720 | 745.836325 |
| 2 | 0.059413 | 0.054502 | -0.088567 | -0.197696 | 0 | 2 | 8.459518 | -2.017247 | 62.606043 |
| 3 | 1.159260 | 2.128179 | -1.251880 | -0.968904 | 0 | 1 | 0.005658 | -5.655181 | -60.851593 |
| 4 | 1.306482 | 0.121526 | -1.995050 | -0.678983 | 3 | 1 | 1.250531 | 1.173030 | 36.743750 |
[3]:
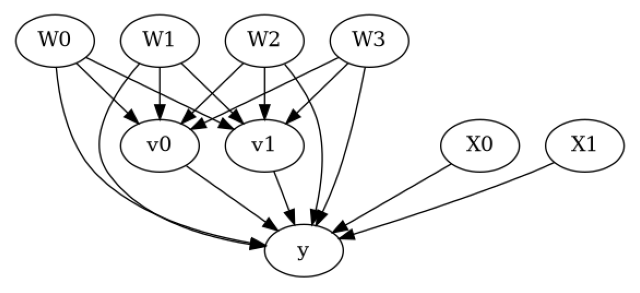
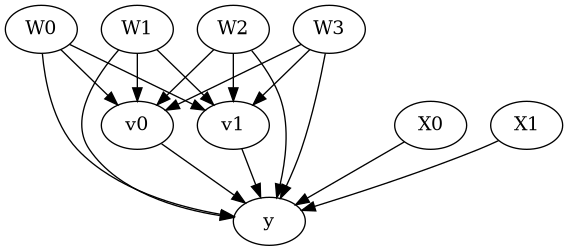
model = CausalModel(data=data["df"],
treatment=data["treatment_name"], outcome=data["outcome_name"],
graph=data["gml_graph"])
[4]:
model.view_model()
from IPython.display import Image, display
display(Image(filename="causal_model.png"))
[5]:
identified_estimand= model.identify_effect(proceed_when_unidentifiable=True)
print(identified_estimand)
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
─────────(E[y|W1,W2,W0,W3])
d[v₀ v₁]
Estimand assumption 1, Unconfoundedness: If U→{v0,v1} and U→y then P(y|v0,v1,W1,W2,W0,W3,U) = P(y|v0,v1,W1,W2,W0,W3)
### Estimand : 2
Estimand name: iv
No such variable(s) found!
### Estimand : 3
Estimand name: frontdoor
No such variable(s) found!
Linear model#
Let us first see an example for a linear model. The control_value and treatment_value can be provided as a tuple/list when the treatment is multi-dimensional.
The interpretation is change in y when v0 and v1 are changed from (0,0) to (1,1).
[6]:
linear_estimate = model.estimate_effect(identified_estimand,
method_name="backdoor.linear_regression",
control_value=(0,0),
treatment_value=(1,1),
method_params={'need_conditional_estimates': False})
print(linear_estimate)
*** Causal Estimate ***
## Identified estimand
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
─────────(E[y|W1,W2,W0,W3])
d[v₀ v₁]
Estimand assumption 1, Unconfoundedness: If U→{v0,v1} and U→y then P(y|v0,v1,W1,W2,W0,W3,U) = P(y|v0,v1,W1,W2,W0,W3)
## Realized estimand
b: y~v0+v1+W1+W2+W0+W3+v0*X0+v0*X1+v1*X0+v1*X1
Target units: ate
## Estimate
Mean value: 49.86459109879479
You can estimate conditional effects, based on effect modifiers.
[7]:
linear_estimate = model.estimate_effect(identified_estimand,
method_name="backdoor.linear_regression",
control_value=(0,0),
treatment_value=(1,1))
print(linear_estimate)
*** Causal Estimate ***
## Identified estimand
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
─────────(E[y|W1,W2,W0,W3])
d[v₀ v₁]
Estimand assumption 1, Unconfoundedness: If U→{v0,v1} and U→y then P(y|v0,v1,W1,W2,W0,W3,U) = P(y|v0,v1,W1,W2,W0,W3)
## Realized estimand
b: y~v0+v1+W1+W2+W0+W3+v0*X0+v0*X1+v1*X0+v1*X1
Target units:
## Estimate
Mean value: 49.86459109879479
### Conditional Estimates
__categorical__X0 __categorical__X1
(-3.252, -0.0735] (-3.1799999999999997, -0.296] -5.959011
(-0.296, 0.302] -3.445913
(0.302, 0.826] -3.262347
(0.826, 1.409] -4.914089
(1.409, 4.772] -1.098686
(-0.0735, 0.52] (-3.1799999999999997, -0.296] 27.392269
(-0.296, 0.302] 28.838159
(0.302, 0.826] 30.072040
(0.826, 1.409] 30.641640
(1.409, 4.772] 32.283592
(0.52, 1.029] (-3.1799999999999997, -0.296] 48.168435
(-0.296, 0.302] 49.191962
(0.302, 0.826] 49.943283
(0.826, 1.409] 51.100576
(1.409, 4.772] 52.287870
(1.029, 1.599] (-3.1799999999999997, -0.296] 68.254575
(-0.296, 0.302] 69.782471
(0.302, 0.826] 70.859518
(0.826, 1.409] 70.892093
(1.409, 4.772] 72.500243
(1.599, 4.389] (-3.1799999999999997, -0.296] 100.358743
(-0.296, 0.302] 102.224670
(0.302, 0.826] 103.162188
(0.826, 1.409] 102.369651
(1.409, 4.772] 104.967850
dtype: float64
More methods#
You can also use methods from EconML or CausalML libraries that support multiple treatments. You can look at examples from the conditional effect notebook: https://py-why.github.io/dowhy/example_notebooks/dowhy-conditional-treatment-effects.html
Propensity-based methods do not support multiple treatments currently.