Demo for the DoWhy causal API#
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
from dowhy.graph import build_graph_from_str
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
nx_graph = build_graph_from_str(data["dot_graph"])
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | 1.734861 | True | 5.677652 |
| 1 | 1.692967 | True | 5.345768 |
| 2 | -0.678012 | True | 5.865227 |
| 3 | 0.320417 | False | -1.465444 |
| 4 | -1.743889 | True | 5.025204 |
| ... | ... | ... | ... |
| 995 | -1.195709 | False | -0.097315 |
| 996 | 1.086976 | False | 1.035984 |
| 997 | 0.115227 | True | 4.093856 |
| 998 | 0.986937 | True | 4.631330 |
| 999 | 0.226167 | True | 5.485148 |
1000 rows × 3 columns
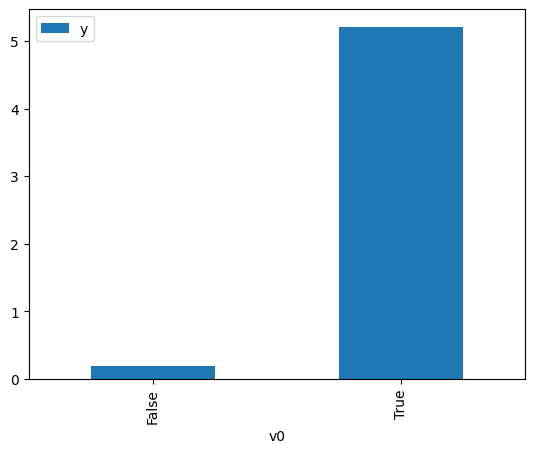
[3]:
# data['df'] is just a regular pandas.DataFrame
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<Axes: xlabel='v0'>
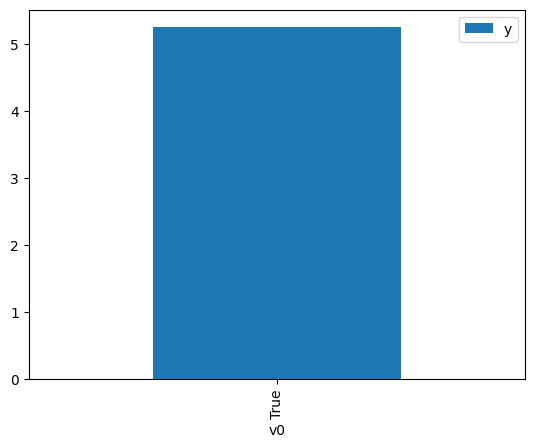
[4]:
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause]
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<Axes: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 1.425122 | False | 1.553599 | 0.468980 | 2.132286 |
| 1 | 1.522968 | False | 2.940058 | 0.464910 | 2.150952 |
| 2 | 1.624560 | False | -0.714125 | 0.460690 | 2.170659 |
| 3 | 1.883998 | False | -1.628021 | 0.449938 | 2.222528 |
| 4 | 0.781901 | False | -1.317539 | 0.495812 | 2.016892 |
| ... | ... | ... | ... | ... | ... |
| 995 | 1.883998 | False | -1.628021 | 0.449938 | 2.222528 |
| 996 | 1.624560 | False | -0.714125 | 0.460690 | 2.170659 |
| 997 | 0.027170 | False | 1.790618 | 0.527316 | 1.896398 |
| 998 | 0.538403 | False | -1.014590 | 0.505985 | 1.976345 |
| 999 | 0.007696 | False | 1.750373 | 0.528127 | 1.893485 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 1.628855 | True | 5.879210 | 0.539489 | 1.853607 |
| 1 | 0.800083 | True | 5.501920 | 0.504947 | 1.980405 |
| 2 | 1.329587 | True | 6.044962 | 0.527042 | 1.897382 |
| 3 | 2.467841 | True | 2.798915 | 0.574072 | 1.741940 |
| 4 | 0.387281 | True | 3.828899 | 0.487704 | 2.050424 |
| ... | ... | ... | ... | ... | ... |
| 995 | 2.490357 | True | 6.209058 | 0.574992 | 1.739154 |
| 996 | 0.802514 | True | 5.794019 | 0.505049 | 1.980007 |
| 997 | 0.835262 | True | 4.489131 | 0.506417 | 1.974659 |
| 998 | 0.080196 | True | 4.995161 | 0.474894 | 2.105735 |
| 999 | 1.739403 | True | 3.959105 | 0.544075 | 1.837982 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression#
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 5.01130335938293$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.0886036474860154$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.934 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.934 |
| Method: | Least Squares | F-statistic: | 7086. |
| Date: | Sat, 12 Jul 2025 | Prob (F-statistic): | 0.00 |
| Time: | 13:59:10 | Log-Likelihood: | -1425.1 |
| No. Observations: | 1000 | AIC: | 2854. |
| Df Residuals: | 998 | BIC: | 2864. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 0.2809 | 0.028 | 9.906 | 0.000 | 0.225 | 0.337 |
| x2 | 5.0450 | 0.052 | 97.125 | 0.000 | 4.943 | 5.147 |
| Omnibus: | 1.607 | Durbin-Watson: | 2.017 |
|---|---|---|---|
| Prob(Omnibus): | 0.448 | Jarque-Bera (JB): | 1.600 |
| Skew: | 0.047 | Prob(JB): | 0.449 |
| Kurtosis: | 2.828 | Cond. No. | 2.33 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.